Many of you know that the precursor to BioGPS was another web tool called SymAtlas. While SymAtlas was great, BioGPS was meant to expand on that design by focusing on community extensibility and user customizability. We are very proud that BioGPS handled all of the key SymAtlas use cases…
… with one notable exception, that is. BioGPS lacked the ability to search for genes by expression pattern, a commonly-used feature in SymAtlas. And our users let us know over and over and over and over again that we needed to add this feature before officially retiring SymAtlas.
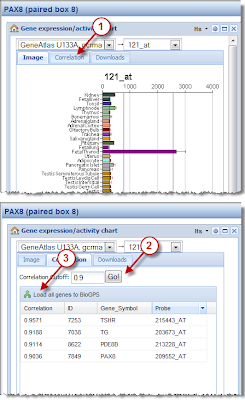
We’re happy to report now that search by correlation is now an official feature in BioGPS. Follow the three steps shown at the right to see how it works. As always, let us know if you have any feedback on this new feature.

Thanks for making this feature available (I was one of those who had been asking…)
I don't want to appear ungrateful, but there are a few additional features I would find useful:
1) At the moment, I don't see a way to save the result of a correlation search on my computer (other than a screenshot)
2) There is an option to save the correlated genes in the current gene list. This feature is useful but could be enhanced by offering a selection which genes to put in the list.
3) This 'save to genelist' feature does not really help to solve problem (1), because the order of the genes is not maintained (are the genes in the gene list sorted by anything in particular?) and the correlation coefficients are lost as well.
Anyway, even in the current state this new feature is very useful and helps with the transition from Symatlas
Suicyte,
First off, sorry for the delayed response due to the Thanksgiving holiday.
1) You're right, we should add an "Export to CSV" button to the correlation table. I've created a ticket to get that added.
2) We can look into adding this feature. In the mean time, you can use this work-around: click the "Load all genes" button, then select the genes you want to save, then click the save button, the load your saved list. If this workflow isn't clear, let me know and I will create a screencast.
3) More intelligent ordering of the current gene list is a ticket we've had on our list for a while now. We're not exactly sure how/if we can save the correlation coefficients themselves though in the gene list, but we'll think about it more.
Thanks for your feedback. It is very much appreciated.
Cheers,
-andrew
correlation search is nice, but i would also like to select tissues of interest as a first pass as was possible in SymAtlas–for example, i would like to query all genes that have expression >3x median in liver, macrophages, and bone. i cannot find this on bioGPS. is it available?
Hi Josie, thanks for your comment. You're right, SymAtlas had two methods of searching by the gene expression patterns — "Search by correlation" and "Search by expression". We were planning on only implementing one, and search by correlation was overwhelmingly more popular. I hope you're able to find a "seed gene" with the expression pattern you want to find.
(… and we've also noted that you would like the old "search by expression" function back. If we hear enough similar feedback, we'll consider it…)
Cheers,
-andrew
The old "search by expression" feature was fantastic. Is it coming back? Why get rid of it? Is there another website that hosts this sort of feature. I'm freaking out that it isn't available anymore. Those of us without NIH budgets used it to identify candidate genes of interest. What is "correlation search". Correlated with what?
Hi Anonymous,
At this moment, we are not planning to implement the "search by expression" feature in BioGPS. We're always open to being swayed by our users though, and your vote here has been duly noted.
There are two alternate solutions I'd propose to you. First, "search by correlation" allows you take a given gene of interest and find other genes that have a similar pattern. So, if you used to use "search by expression" to find liver-expressed genes, now you might search first for CYP1A2, a gene that is commonly known to be highly expressed in liver. From there, you can use the "search by correlation" function as described in this blog post.
Second, you can download the raw data on our downloads page. With the files there, you can do some basic filtering in excel, and with a small amount of work with formulas you can do the exact same queries as the old "search by expression". Or, you can take the data file to your local bioinformatician for more help.
Hope that helps…
-andrew
Hi
I wanted to ask that can we use the normalized gene atlas data and directly do the Pearson coreelation or we have to again normalize my taking the mean for each gene?
Hi Amitabh,
I'm not sure if I'm interpreting your question exactly right, but there should be no difference in the Pearson correlation if you first do mean-centering versus using the raw values. The Pearson calculation already takes in account a mean subtraction. Please post again with more details if you need more information.
Cheers,
-andrew
Correlation and dependence are any of a broad class of statistical relationships between two or more random variables or observed data values. perason correlation