 In our previous post, we described how we created a prototype Guesser program that plays the game 20 Questions on genes. The next natural question is: how accurate is our guesser?
In our previous post, we described how we created a prototype Guesser program that plays the game 20 Questions on genes. The next natural question is: how accurate is our guesser?
To assess accuracy, we created an evaluation framework that serves as as 20 questions playing partner — the Answerer. The Answerer randomly selects a gene, and then it retrieves all known Gene Ontology annotations for that gene. The Answerer is then set up to play the 20 Questions game with the Guesser, essentially providing answers for the questions it is asked. If the Guesser guesses the random gene in 20 questions or less, then it wins. And then we repeat this exercise 100 times.
Using this approach, we find that our guesser has a 100% accuracy, and correctly identifies the gene in an average of 10 guesses. Not bad…
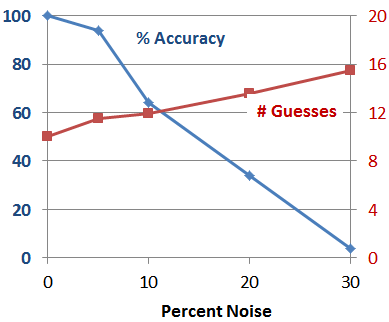
To challenge our guesser program a little bit more, we injected variable amounts of noise in the Answerer’s responses. Not surprisingly, as we increase the percentage of incorrect answers, the success rate goes down while the average number of guesses required for each success went up.
This trend is even more stark when plotting these statistics relative to the number of incorrect answers given on a gene-by-gene basis (since even in the 30% error matrix, there are some trials in which every answer given by the Answerer was correct):
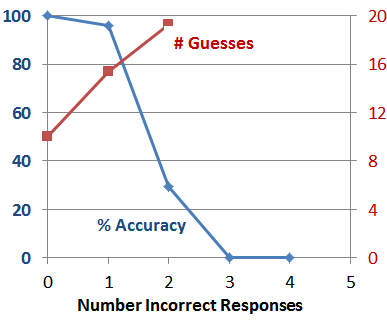
So what does this all mean? The good news is that our prototype Guesser is pretty good as long as the Answerer’s answers pretty much agree with gene annotations in the databases. The bad news is that the gene annotations in the source databases are undoubtedly incomplete relative to what is actually known by experts on those genes. Which is why our Guesser is rarely successful when playing with a human Answerer.
This gap between the community’s knowledge of gene function and the knowledge represented in structured annotation databases is both the challenge and the opportunity. The challenge is to write a better Guesser that is more robust to errors in the database. The opportunity comes from the ability to add knowledge in structured databases from “incorrect” responses from humans. Those “incorrect” responses will usually be bits of knowledge that just haven’t made it into structured GO annotations, and we think our game can be a useful tool to close the gene annotation gap.
(For those who are interested, the R code to do the evaluation can be found in the 20-evaluation code repository…)
