 Through the generous support from NIGMS, we have enjoyed stable funding for BioGPS development since 2008. During that time, we think we’ve made great progress building our community-extensible and user-customizable gene portal.
Through the generous support from NIGMS, we have enjoyed stable funding for BioGPS development since 2008. During that time, we think we’ve made great progress building our community-extensible and user-customizable gene portal.
It’s worth recapping how the last four years have gone relative to what we originally proposed. For completeness, I’m posting the full text of the proposal, but let me summarize the four aims and what we’ve accomplished:
Specific Aim #1: Incorporate community-generated data by allowing users to upload custom numeric data sets for analysis and visualization.
- What we proposed: Many users love BioGPS for the simple bar chart expression viewer coupled with a simple gene search interface. (We like simplicity.) Many users have emailed over the years asking to view other data sets through BioGPS, including their published and unpublished data.
- What we did: This aim has taken the longest to get just right, but I’m happy to announce that we’re very close to releasing a significant new release here. In short, we’re going to be increasing the number of data sets available through BioGPS by an order of magnitude or two, plus we’re setting the stage for more increases after that. More details soon.
Specific Aim #2: Incorporate community-generated gene annotation by seeding a “gene wiki” with structured gene portal content.
- What we proposed: We envisioned a “Gene Wiki” within Wikipedia whose goal was to produce a collaboratively-written, community-reviewed, and continuously-updated review article for every human gene.
- What we did: We tackled this work early in the grant period, and it has blossomed more than we could have imagined at the outset. The Gene Wiki is now the subject of its own grant, and it has been the subject of many recent publications.
Specific Aim #3: Incorporate community-generated plugins by creating simple programmatic interfaces for external developers to extend BioGPS functionality.
- What we proposed: To embrace the principle of community-extensibility, we designed a plugin architecture that allows any user to add new content to BioGPS without any involvement or delay from our developer staff.
- What we did: This plugin interface in many ways is the core of the BioGPS architecture. By generalizing online gene-centric resources by a URL template, we have created the most comprehensive index of web pages on gene properties that currently spans over 250 unique resources.
Specific Aim #4: Enable users to share and customize the usage and layout of BioGPS plugins through optional user accounts.
- What we proposed: We designed a powerful and customizable layout system that would allow users to mix and match gene annotation resources according to their individual use cases.
- What we did: We provided several default layouts that cover common use cases, from literature searching, to accessing orthologs in model organism databases, to consulting pathway resources. In addition, BioGPS users have created over 2000 custom layouts according to their individual needs.
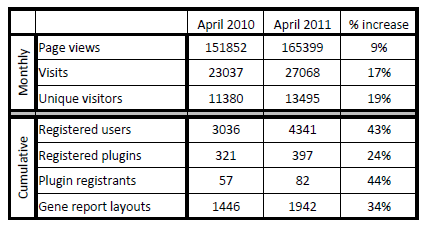
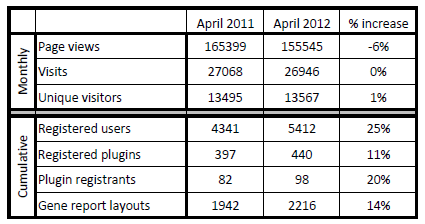
And I realize it’s been a while since we’ve disclosed our BioGPS usage statistics. I’m posting two tables, one from last year’s progress report comparing 2011 to 2010, and one that will be included in this year’s progress report:
Well, it looks like we’ve plateaued a bit on the user base. I chock that up to our lab’s transition to Scripps in the last year (and the inherent difficulties keeping up double-digit growth). But we’re redoubling our efforts to keep growing BioGPS, and I think our upcoming data set features will go a long way to expanding our user base even further.

Trackbacks/Pingbacks