This is the fourth blog post in a series on our Gene Wiki renewal. More details below.
Our plans to build a Centralized Model Organism Database (CMOD) is the one that we discussed most openly as we were drafting our proposal. So you should feel free to read our previous blog post on this subject, but this post is a slightly expanded and refined version.
The Gene Wiki is a crowdsourcing mechanism to create gene-specific review articles. While these human-readable articles are fantastic resources, they are not easily amenable to computer-readability and bioinformatics analyses. CMOD is a parallel initiative to organize structured gene annotations via crowdsourcing. This aim will take advantage of broader initiatives to crowdsource structured data (specifically, Wikidata and/or Freebase).
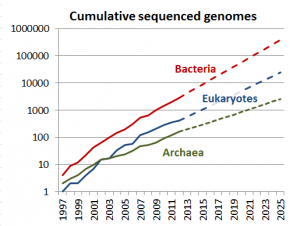 To be clear, CMOD is in no way a replacement for the amazing work done by professional curators. But, we as a community need to recognize that those professional curation efforts simply do not scale with the exponential growth in genome sequencing and genomic science. As a thought experiment, how many model organism databases can you name? If you can get to 10 you’re doing pretty well, and I think 30 would be a stretch for most scientists. Regardless what number you reach, CMOD is not aimed at those organisms, but rather the other ~8000 sequenced species (and growing) for which no biocuration resources exist.
To be clear, CMOD is in no way a replacement for the amazing work done by professional curators. But, we as a community need to recognize that those professional curation efforts simply do not scale with the exponential growth in genome sequencing and genomic science. As a thought experiment, how many model organism databases can you name? If you can get to 10 you’re doing pretty well, and I think 30 would be a stretch for most scientists. Regardless what number you reach, CMOD is not aimed at those organisms, but rather the other ~8000 sequenced species (and growing) for which no biocuration resources exist.
There are two types of annotations that CMOD will handle — gene and genome annotations.
Genome annotation refers to the description of genomic features and their coordinates. These features include genes, exons, promoters, and operons. For example, NCBI, Ensembl Bacteria, and PATRIC are three of the largest and most systematic providers of microbial genome annotations, collectively annotating over 100 million features on thousands of sequenced microbial genomes.
While these resources are incredibly valuable, they are by no means complete. Over 97% of all features relate to the gene boundaries themselves (‘gene’, ‘CDS’, ‘exon’). In contrast, only 57 of the 2515 bacterial species at NCBI had any annotated operon, promoter, attenuator, regulatory region, or terminator feature (0.1% of all features), and no such annotations were found in Ensembl Bacteria or PATRIC.
As a case in point, the transcriptional regulation of the Listeria monocytogenes genome was extensively characterized in a prominent Nature paper1, revealing (among other findings) the existence of 517 operons and 103 small regulatory RNAs. However, those data are not visible through any genome browser, nor are they available for download at any of the data repositories examined. In fact, those data are currently only available in PDF format as supplementary info on the journal website.
The lack of accessibility of these published data is a failure of the scientific publishing system. Clearly CMOD won’t fix that problem at its root, but it will vastly improve the situation since one scientist can structure that data, deposit it into our open community-based database, and then that data can be freely accessed and queried by the rest of the research community.
Gene annotations refer to the description of genes and their protein products, and most often, gene annotations are based on the Gene Ontology (GO). Among bacterial species, Mycobacterium tuberculosis and Escherichia coli are by far the most well-annotated species, each with over 10,000 experimentally-derived gene annotations. Overall, there are 189 bacterial species with at least five such annotations, and in total, there are ~55,000 experimentally-derived, bacterial GO annotations.
While these existing annotations are valuable, we hypothesized that they are also highly incomplete. To estimate the total number of species that could have GO annotations, took all entries in NCBI Taxonomy and searched the species name with “gene” in PubMed. We found 1,107 species that returned at least 10 PubMed hits. Again to highlight an extreme example, consider Borrelia burgdorferi, the bacterial species that causes Lyme Disease whose genome was sequenced in 19972. Searching PubMed for “Borrelia burgdorferi” AND gene reveals over 1,400 articles. However, there are no GO annotations for any of the ~1,300 coding sequences, despite the discovery of many functional roles for genes in the literature3.
So based on our goal of crowdsourcing gene and genome annotations, we broke this aim down into three parts:
- Develop tools to import genome annotation data from GFF3 files to Wikidata. These tools will be use to import all existing genome annotations from genomics resources. This step will bring together all existing data in one framework, and also open it up to additions and edits by the community at large, which is especially important for species without a dedicated model organism database.
- Adapt the JBrowse genome browser and WEb Apollo annotation editor to use the Wikidata API. A genome browser is the most basic and essential visualization of genome annotation data, and modified versions of these popular tools will mean that any changes to CMOD genome annotations will be immediately visible to the rest of the scientific community.
- Develop tools to import gene annotation data from GAF files to Wikidata. Analogous to the first part of this aim, this step will ensure that existing gene annotation knowledge will be available within CMOD. Building on that foundation, the entire scientific community will be able to add and edit annotations using Wikidata interfaces.
In the face of exponential growth of genome sequencing (including systematic efforts to preserve the Earth’s genomic biodiversity), we think crowdsourcing and CMOD are resources that will be instrumental for future genomic research.
This blog post is part of a series of entries on our NIH proposal to continue developing the Gene Wiki. The other posts are here:
Post #0: Introduction
Post #1: Gene Wiki progress report
Post #2: Aim 1: Diseases and drugs
Post #3: Aim 2: Outreach
Post #4: Aim 3: Centralized Model Organism Database (this post)
Post #5: Aim 4: Patient-aligned crowdsourcing

This is a fabulous idea. What are your criteria for model species? A sequenced genome?
@Colleen, in theory, any species with at least some minimal amount of assembled genomic sequence OR some minimal amount of GO annotations for specific genes would be suitable. That admittedly casts a pretty broad net, but I think that underscores how unmet this particular need is…