This is the fifth and final blog post in a series on our Gene Wiki renewal. More details below.
Crowdsourcing is all about motivating large groups of individuals to collaboratively achieve some shared vision. For example, the Wikipedia crowd primarily uses volunteerism as a motivation. Other recent initiatives motivate crowds of gamers using fun and enjoyment1.
 In this aim, we will target a crowd of “patient-aligned individuals”. So many people have been personally impacted by serious diseases, either themselves personally or one of their loved ones. Our goal is to create a mechanism by which these individuals can directly contribute to biomedical research.
In this aim, we will target a crowd of “patient-aligned individuals”. So many people have been personally impacted by serious diseases, either themselves personally or one of their loved ones. Our goal is to create a mechanism by which these individuals can directly contribute to biomedical research.
We believe that this community would enthusiastically welcome that opportunity to contribute. This point was made especially clearly by our recent Twitter interactions that showed how active and engaged patients have become in their own health care and research. This is especially true for the so-called “rare diseases” collectively affect over 25 million people in the United States alone for which drugs are often not available.
How do we want to empower non-scientists to impact biomedical research? Our application will address the challenge of annotating the massive biomedical literature. Consider that there are over one million new research articles published every year, roughly equivalent to one article every thirty seconds.
Keeping up with the literature is drinking from the proverbial fire hose, and every researcher I know struggles with that challenge. It is difficult to identify the small percentage of articles that is relevant to my research, much less to extract the knowledge that they contain and put it in context of past research. And time matters as well — the longer it takes for me to discover the latest relevant research, the longer it takes for me to incorporate those new findings into my own research.
We want to enable non-scientists to help us “annotate” the latest research articles immediately as they are published. Annotations can be separated into two distinct types. First, “entity recognition” refers to the process of highlighting all the biomedical concepts, most notably the diseases, genes, and drugs mentioned in an article. Systematically annotating the concepts in every article benefits scientists on a number of levels. Consider for example a chordoma researcher. That researcher could be notified when a new article on chordoma is published. Or when any article mentions chordoma with a drug. Or chordoma and the T gene.
The second step in the annotation process involves “relationship extraction” to define which of those concepts identified in step one are related to each other and how. For example, we want to annotate that the microRNA miR-31 is related to chordoma because of its downregulation in tumors, and rapamycin is related to chordoma as an “inhibitor of chordoma cell proliferation” and for its activity “reducing the growth of chordoma xenografts”. Aggregating all of these annotated relationships into a single database would result in a powerful resource for researchers for many downstream applications.
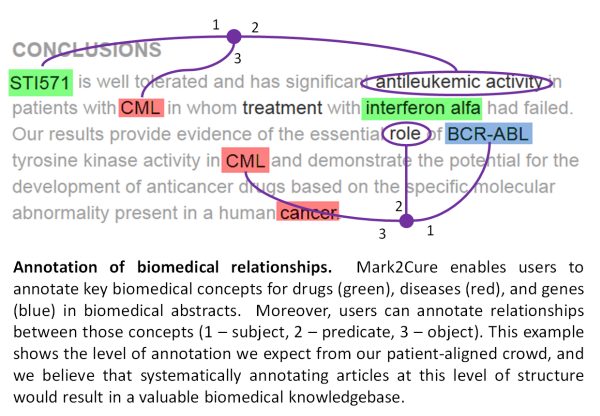
Importantly, we don’t think non-scientists need to understand the scientific meaning of these biomedical concepts and relationships (though we certainly don’t rule it out either). Rather, we think that we can train users to use their basic English language skills to learn to recognize disease, genes, and drugs (and to use online resources to confirm their guesses), and to use the rules of grammar to simply highlight the phrase that describes the interaction.
We are calling our application Mark2Cure. If you are interested in getting notified when we launch, please sign up to our email list at mark2cure.org.
This blog post is part of a series of entries on our NIH proposal to continue developing the Gene Wiki. The other posts are here:
Post #0: Introduction
Post #1: Gene Wiki progress report
Post #2: Aim 1: Diseases and drugs
Post #3: Aim 2: Outreach
Post #4: Aim 3: Centralized Model Organism Database
Post #5: Aim 4: Patient-aligned crowdsourcing (this post)

There are several problems with the general model that you are proposing/implementing. First, you don’t need a faceless Crowd to scale paper analysis. The authors know a lot more about the papers than any reader, and in most cases are still with us, and don’t like to be mis-quoted. Why not just ask the authors to annotate their papers? (In most cases the corresponding author’s email is automatically available, so this could easily be automatically done). A hybrid model would be to allow the authors to repair Crowd-based annotations.
Secondly, one will almost always want to be able to cross-annotate across papers, and more importantly, from papers to a variety of DBs, making reference from one annotation of the sort you describe to others. The backing wiki technology may help with this a little, but it sort of defeats your elegant annotation model. (Of course, the backing semantic engine is all about this, but that’s for computers, not people — some “smarts”, however, could mitigate, like semantic autofill.)
However, all of this is called into question by the third, and far worse difficulty, that papers don’t generally “mean” anything out of context (technical context, as above), and the meaning floats (historical context — by which I don’t mean the socio-cultural history, but the dynamics of the technical semantics); Go, for example, is a very poor functional annotation scheme, and changes, but any such scheme will have this sort of issue (Selfie: http://bioinformatics.oxfordjournals.org/content/19/15/1934.full.pdf). The general point is that a static annotation scheme will fail nearly immediately due to semantic float.
Cheers,
‘Jeff
Jeff, 1st nothing about the idea rules out the idea of inviting authors to annotate their own papers. Undoubtedly some would respond to such a request and some would not. The amount that would not bother as well as the variability in how even a domain expert would conduct the annotations make the ability to tap into a crowd very valuable if achieved.
2nd, cross-annotating papers (establishing links between concepts in different papers), could be an extension of this work. You could imagine an interface that would allow you to find related papers and then do the same kind of arc-building, text selecting etc. across two at once. However, to make real use of this I’d push first on an underlying semantic infrastructure that would unearth those connections automatically based on the knowledge extracted from individual papers. (Or simply use the data to refine NLP models…)
3rd, and this relates to 1., why do you think this would need to be a “static” annotation scheme? If, (and this is the big if here), we identify thousands of people willing to do this kind of work on ongoing basis for fun and for the good of humanity, then there is no reason we could not revisit papers and adapt annotations to account for semantic float. It seems that we would have more of an opportunity to deal with this problem given the large potential workforce than existing annotation centers with their limited human resources.
As you note (and as should be obvious anyway, since I wrote all three in one comment), all three points are related, but I don’t think that the issue of semantic float is as simple as you believe it to be, and having more annotators can actually make it worse, rather than better. Think of this as if you had a large number of programmers hacking the same (or related) pieces of code, trying to put it all together via git (or whatever). There a large number of ways to make this point, but let’s consider just a few:
Example 1. How will you deal with conflicting annotation? (This is the one most obviously exacerbated by having multiple annotators. Some contradictions arise from the papers themselves, but other arise from multiple annotators having differing interpretations. If you don’t believe this will happen, just look at the edit wars that are fought in the change histories of more-or-less any wikipedia page…and I’m sure that, using my git analogy above, you yourself can imagine the cringing pita it would be to have to have ten or a hundred or a thousand hackers all trying to do git merges against you. An infinite number of annotators (programmers) just makes this problem infinitely worse! (If you do it my way, and ask the authors first, you have much less of a problem here. Basically, there’s one final authority. Granted, you have to get their attention, but I think that that’s less hard than getting the attention, and then making sense of, the merges of ten or a hundred randoms.)
Example 2. What happens when the annotation framerwork (e.g., GO) is changed? You may be able to tell what needs to be updated (maybe), but in general you can’t figure out how to update it. This can be as hard (much harder, actually) as the general database versioning problem (or general merge problem, as git), which is unsolved. Many annotators with a great deal of time on their hands, combined with some smart algorithms can help somewhat in simple cases, but not in the general case; consider, for example, the special case where an upper node is changed, and you need to reconsider every assertion in the db — or anyway, every annotation under the changed one, which could be very many of them. Are you going to farm this out, and how do you know w/o got-like version control when the db has stabilized — or even if it will stabilize? There are numerous other problematic special cases of this that I won’t bother spelling out.
The more general point regarding semantic float is that the semantics (and, the point of my paper, cited previously) is that the semantics are, in effect, defined by the annotations, NOT (as most people seem to believe) by GO (or whatever so-called “semantic” backbone things are linked to). You’d be better off, IMHO, binding across papers (with some sort of version control on the assertions) per my second suggestion in the OP, than binding to the GO (etc.) Or, put in a more general way, consider the GO bindings as one case of an inter-document assertion (per my second point in the OP), which no special status over and above any other. If you grant some centralized semantics a special place in the model, when it changes (which it does), you have a mess on your hands. (Another selfie that provides a glimpse of a possibly helpful technical concept in the “bayes community” idea: http://shrager.org/vita/pubs/2009bayescommunity.pdf)
Actually, you’re going to have a mess on your hands regardless. But that’s sorta just the way symbolic biocomputing is, I guess. We muddle through. I just don’t think that depending upon clown sourcing is going to help you muddle through any better, and in many cases, as above, might actually hurt.
Cheers,
‘Jeff
🙂 clown sourcing.. nice (though some clowns may find it offensive). In our experience, the following is not true: “I think that that’s less hard than getting the attention [of an expert on a topic or the author of a paper], and then making sense of, the merges of ten or a hundred randoms”.
I don’t think code repos are a good analogy here. There is no “version”. There is a collection of largely overlapping statements that will be merged and weighted statistically.
Thank you for the reference to the bayes community idea, it looks like it could be very useful. We like a mess.. much better than a vacuum.
I’m curious about how the inputs and outputs to this process will work. Often times researchers have saved PubMed or Google Scholar searches which they use to keep themselves up-to-date. Will researchers be able to register an RSS feed of a PubMed search as an input? Or perhaps a list of PMIDs? Where will the text of the paper come from? Mining PMC in the past has presented some difficulties since the copyright to the article may or may not permit you to mine it.
How will people be able to query and view the results after the annotations have been made? Are you planning on using Cytoscape.js or some similar framework to render the results?
Mark, great questions, thank you. The role of the researcher is up for debate. On one end of the spectrum, we are considering controlling all of the inputs ourselves – basically targeting large, relevant corpuses and providing the research community with the output. On the other end (and I personally lean this way), we are looking at a model of researchers playing very central roles in defining the document collections to be annotated and in specifying the goals of the annotation. Imagine that a researcher could basically issue, promote, organize, and consume the results of an annotation “quest”. I like this model because it distributes hard labor, engages the research community more directly and I think provides a natural pattern for increasing community engagement over time. Your thoughts ?
We currently have a home grown javascript network viewer that shows the results of your own annotation efforts, but I think more purpose-driven applications that consumed the data produced would be ideal. (I’m not a fan of the current use of the network vis, but other team members disagree). And of course the raw data will be available. We would welcome talented app developers to help with these ;)..
Hi Ben,
Thanks for the reply. I think I’m more on the side of researcher-guided approach, since they’re more likely to have an organized corpus to begin with. Also, it would be useful if there were some feedback mechanism so that participants could improve their abilities.
The blog post and tweets started the gears whirring, and it occurred to me that it might be nice to approach some of the researchers in the standup2cancer, or the PanCan organization, since they have access to both the researchers and the patient-aligned crowds that you’d like to tap into.
When you say “the raw data will be available” is that as RDF, or some other format?
Mark, thanks for the suggestions regarding standup2cancer and PanCan. We will add them to the list! We would make the data available in whatever format was most useful to the most developers and would follow any major standards that we identified. Would it make you more likely to hack on if we delivered the results as RDF?