We recently gained access to the anonymised logs of several hundred million SPARQL queries from the Wikidata SPARQL endpoint. This blog post contains some discussion about the main takeaway points, while the full analysis and code can be found here.
What are we looking for?
We are interested in biomedical properties and items in Wikidata, specifically looking at how these entities are used within SPARQL queries, and how often they are searched for. While it would great to look at biomedical entities in the results of SPARQL queries, or being used as intermediates within the knowledge graph, it would be very difficult to do this at this time, and so we are only looking at how items or properties are explicitly used within the SPARQL queries themselves.
Why do we care about this?
Part of the work we do in the Su Lab involves working with data curators and data providers to make their data more findable, accessible, integratable and reusable. Using these query logs, we can get a sense of if we are succeeding. In addition, data providers often have to gather usage metrics to justify additional funding, or make decisions on future improvements to their data. Looking at the query logs can help get a sense of how people are using certain data.
What is in the data?
Please see here for a full description. Briefly, each line in the file dumps contains (1) the anonymized query (reformatted and processed for reducing identifiability), (2) timestamp, (3) source category (robotic or organic, explained here), (4) user agent: A simplified/anonymised version of the user agent string that was used with the request.
How was the data processed?
See here for the main script. Briefly, all 3 files were concatenated together and then the query strings were sorted, grouped and counted. In this way, we could have a set of unique queries, and the number of times each query was executed. We then counted the number of items or properties explicitly mentioned within each query, recording the number of unique queries and total queries each ID was found in. We also then looked at co-occurrence of properties within SPARQL queries. For example, counting the number of times Disease Ontology ID (P699) and subclass of (P279) are used together within the same query.
What did you find??
In this analysis, I’m mostly going to show the unique counts (i.e. if the exact same query is executed many times, it only counts once). The total counts are available with the full analysis.
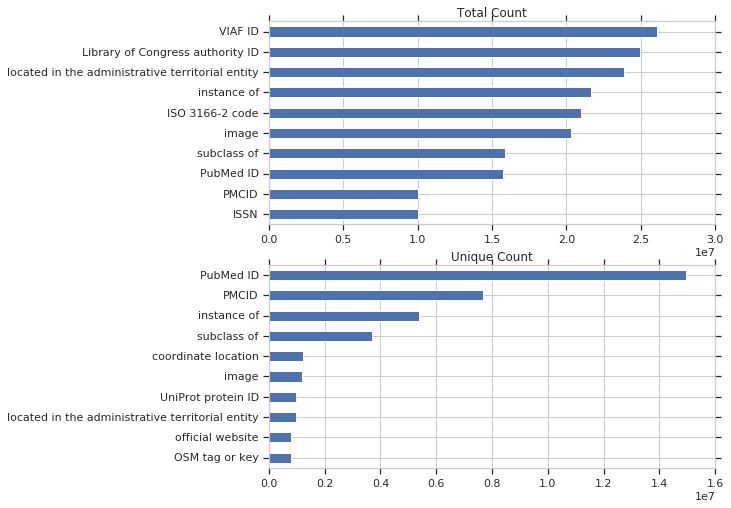
Below are the counts for the most used properties by total query count and by unique query count
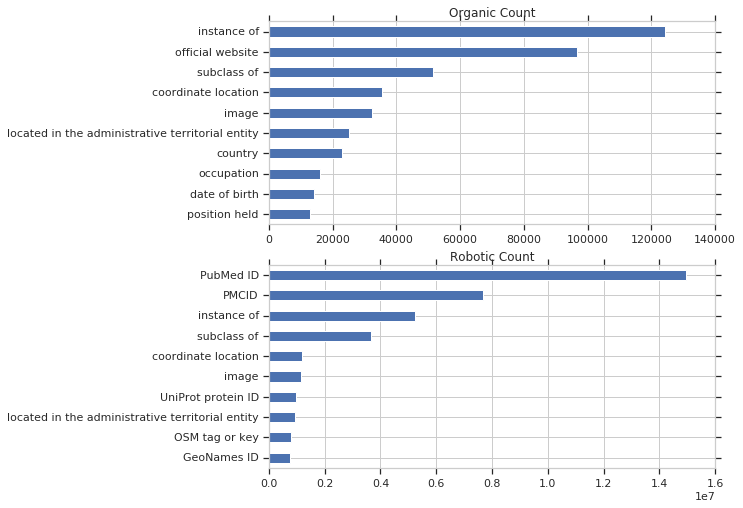
Below, the unique query counts are broken down into organic and robotic query counts. Takeaways for me: The scale is significantly different between organic and robotic queries. As a benchmark, out of 208 million total queries, less than 1 million were classified as organic. The most common unique robotic queries involved PubMed IDs, which make since given the scale of WikiCite, while the most common organic queries uses instance of and subclass of, which make sense given that these are very widespread and useful..
I decided to take a look at the PubMed ID queries, to get an idea of what they are typically. Here are a couple of them:
SELECT * WHERE { ?var1 ( <http://www.wikidata.org/prop/P698> / <http://www.wikidata.org/prop/statement/P698> ) "10000006". }
SELECT * WHERE { ?var1 ( <http://www.wikidata.org/prop/P698> / <http://www.wikidata.org/prop/statement/P698> ) "10000007". }
SELECT * WHERE { ?var1 ( <http://www.wikidata.org/prop/P698> / <http://www.wikidata.org/prop/statement/P698> ) "10000008". }
SELECT * WHERE { ?var1 ( <http://www.wikidata.org/prop/P698> / <http://www.wikidata.org/prop/statement/P698> ) "10000009". }
As you can see, many of the queries are the same query with different IDs.
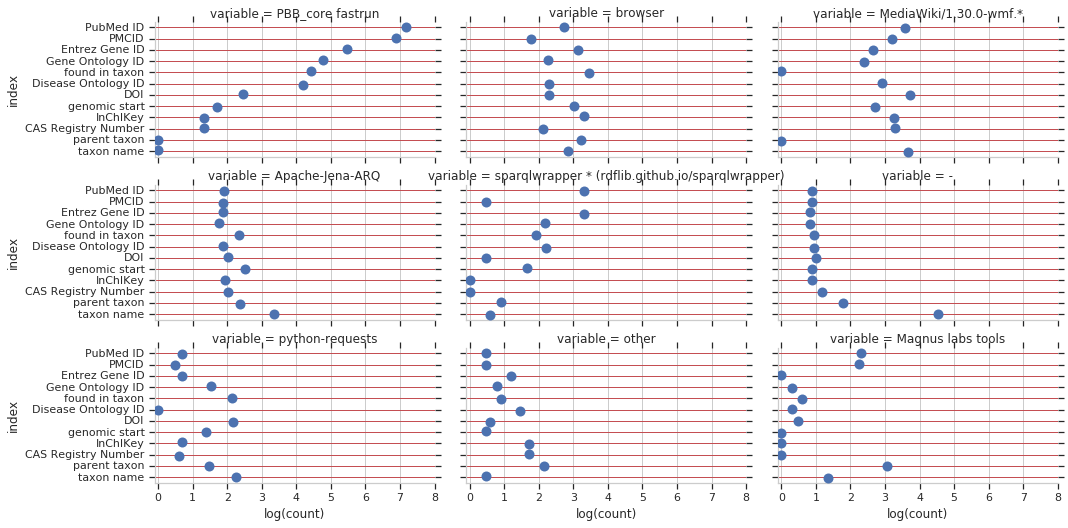
Next, I looked at property usage stratified by useragent, for both unique and total query counts.
For fun, I was also curious what the most queried query was. It is the query below, which is looking for MusicBrainzIDs. It was executed 9,721,509 times! As the strings were replaced by “string1”, “string2”, etc. it is probably the case that this query was executed with different strings each time, but those have been anonymized. The other top 10 can be seen on github.
SELECT ?var1 ?var2 WHERE { VALUES ( ?var2 ) { ( “string1” ) ( “string2” ) ( “string3” ) ( “string4” ) ( “string5” ) ( “string6” ) ( “string7” ) ( “string8” ) ( “string9” ) ( “string10” ) ( “string11” ) ( “string12” ) ( “string13” ) ( “string14” ) ( “string15” ) ( “string16” ) ( “string17” ) ( “string18” ) ( “string19” ) ( “string20” ) ( “string21” ) ( “string22” ) ( “string23” ) ( “string24” ) ( “string25” ) ( “string26” ) ( “string27” ) ( “string28” ) ( “string29” ) ( “string30” ) } ?var1 <http://www.wikidata.org/prop/direct/P434> ?var2 . }
Lets look at biomedical properties!
As you may know, the at the Su Lab we have a project where we enrich Wikidata with biomedical data. See here and here for more info. Part of this involves working with data curators and data providers to make their data more usable by the Wikidata community. Using these query logs, we can get a sense of if we are succeeding in making these biomedical data findable, accessible, and integratable through Wikidata.
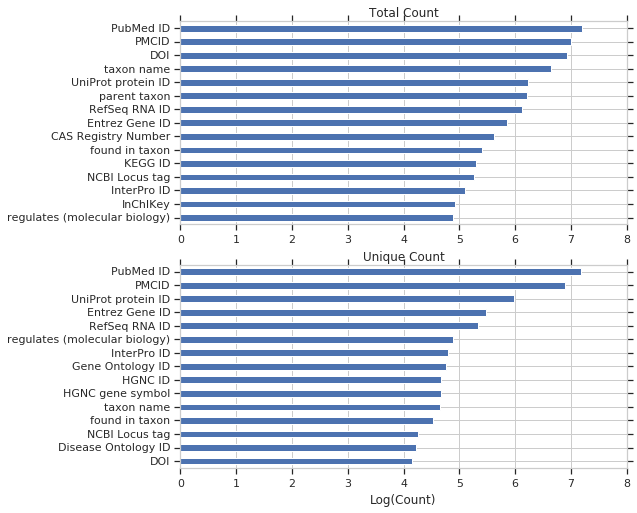
We’ll start with biomedical properties, showing the unique and total query counts.
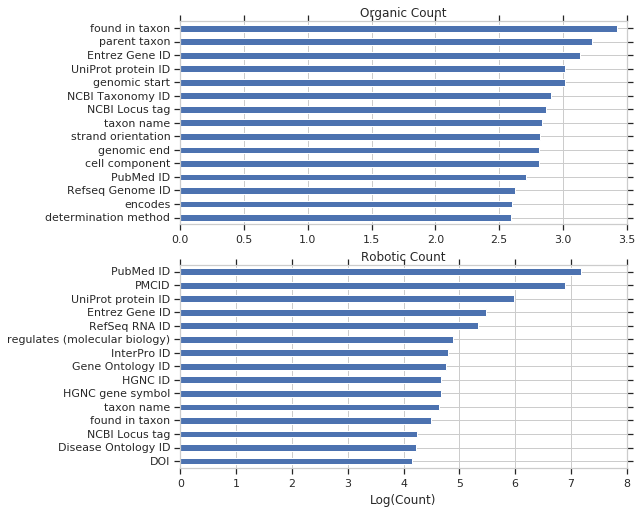
And broken down into organic and robotic query counts.
And lastly, property usage stratified by useragent and property. Takeaways for me: PBB_core (aka WikidataIntegrator) and Magnus’s tools are the largest queriers for reference items. Taxons, drugs, diseases, and genes are used highly through MediaWiki and through browser-based queries.
In the next blog post I’ll talk about item usage in queries, and co-occurrence of properties within the same query.